It seems people just can’t resist playing God. I know I love making human-animal hybrids, intelligent robot slaves, and generally changing the size of animals for fun. And now some researchers have gone and tried to expand the number of bases of DNA.
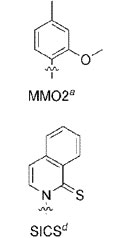
First of all, the new bases, D and M (dSICS and dMMO2), have the stupidest names imaginable. Someone rename them things like damnedinine and monsterine quick! A bunch of letters is unacceptable. Secondly, we’ve had other bases pair up for years. RNA is rip with this, it is not that new. Thirdly, none of these are coded into any codon triplets yet. So this is basically the equivalent of announcing you’ve invented two new letters for the alphabet but haven’t invented any words to use them in. Color me unimpresses. Once you got working super-rats that have thirteen arms thanks to their monsterine bases coding for Tyro-mega-lysine Amino Acid thank you will have my interest. Until then, go suck on a pipetter!
Artificial letters added to life’s alphabet
* 13:07 30 January 2008
* NewScientist.com news service
* Robert AdlerTwo artificial DNA “letters” that are accurately and efficiently replicated by a natural enzyme have been created by US researchers. Adding the two artificial building blocks to the four that naturally comprise DNA could allow wildly different kinds of genetic engineering, they say.
Eventually, the researchers say, they may be able to add them into the genetic code of living organisms.
The diversity of life on earth evolved using genetic code made from arrangements of four genetic “bases”, sometimes described as letters. They are divided into two pairs, which bond together from opposite strands of a DNA molecule to form the rungs of its characteristic double-helix shape.
The unnatural but functional new base pair is the fruit of nearly a decade of research by chemical biologist Floyd Romesberg, at the Scripps Research Institute, La Jolla, California, US.
Romesberg and colleagues painstakingly created a library of nearly 200 potential new genetic bases that are slight variations on the natural ones. Unfortunately, none of them were similar enough in structure and chemistry to the real thing to be copied accurately by the polymerase enzymes that replicate DNA inside cells.
Random generationFrustrated by the slow pace designing and synthesising potential new bases one at a time, Romesberg borrowed some tricks from drug development companies. The resulting large scale experiments generated many potential bases at random, which were then screened to see if they would be treated normally by a polymerase enzyme.
With the help of graduate student Aaron Leconte, the group synthesized and screened 3600 candidates. Two different screening approaches turned up the same pair of molecules, called dSICS and dMMO2.
The molecular pair that worked surprised Romesberg. “We got it and said, ‘Wow!’ It would have been very difficult to have designed that pair rationally.”
But the team still faced a challenge. The dSICS base paired with itself more readily than with its intended partner, so the group made minor chemical tweaks until the new compounds behaved properly.
Novel DNA“We probably made 15 modifications,” says Romesberg, “and 14 made it worse.” Sticking a carbon atom attached to three hydrogen atoms onto the side of dSICS, changing it to d5SICS, finally solved the problem. “We now have an unnatural base pair that’s efficiently replicated and doesn’t need an unnatural polymerase,” says Romesberg. “It’s staring to behave like a real base pair.”
The team is now eager to find out just what makes it work. “We still don’t have a detailed understanding of how replication happens,” says Romesberg. “Now that we have an unnatural base pair, we are continuing experiments to understand it better.”
In the near future, Romesberg expects the new base pairs will be used to synthesize DNA with novel and unnatural properties. These might include highly specific primers for DNA amplification; tags for materials, such as explosives, that could be detected without risk of contamination from natural DNA; and building novel DNA-based nanomaterials.
Increased ‘evolvability’More generally, Romesberg notes that DNA and RNA are now being used for hundreds of purposes: for example, to build complex shapes, build complex nanostructures, silence disease genes, or even perform calculations. A new, unnatural, base pair could multiply and diversify these applications.
The most challenging goal, says Romesberg, will be to incorporate unnatural base pairs into the genetic code of organisms. “We want to import these into a cell, study RNA trafficking, and in the longest term, expand the genetic code and ‘evolvability’ of an organism.”
Stanford University chemist Eric Kool, has studied the fundamental chemistry of base-pair bonding. He foresees challenges, but great potential in the unnatural bases.
“It requires a long effort by multiple laboratories, but I think ultimately it will lead to some important tools,” he says. “The ability to encode amino acids with unnatural base pairs will be quite powerful when it comes.”
Journal reference: Journal of the American Chemical Society (DOI: 10.1021/ja078223d)
The new bases in question: